Keywords:
Epigenetics, oxidized pyrimidine bases, mass spectrometry, proteomics.
Abstract:
Epigenetic information is stored in the form of modified bases in the genome. The positions and the kind of the base modifications determines the identity of the corresponding cell. Setting and erasing of epigenetic imprints controls the complete development process starting from an omnipotent stem cells and ending with an adult specialized cell. I am going to discuss the latest results related to the function and distribution of the epigenetic marker bases 5-hydroxymethylcytosine (hmC), 5-formylcytosine (fC), 5-carboxycytosine (caC) and 5-hydroxymethyluracil (Scheme 1).[1] These nucleobases control epigenetic programming of stem cells and some of these bases are also detected at relatively high levels in brain tissues. Synthetic routes to these new bases will be discussed that enable today preparation of oligonucleotides containing the new bases. The second part of the lecture will cover mass spectroscopic approaches to decipher the biological functions of the new epigenetic bases of which some were described in the past as pure DNA lesions.[2] In particular, results from quantitative mass spectrometry, new covalent-capture proteomics mass spectrometry and isotope tracing techniques will be reported. The data allow us to unravel the chemistry in stem cells and the protein networks that are controlled by the epigenetic base modifications.[3] Finally I am dicussing potential präbiotic origins of these modfied bases.[4]
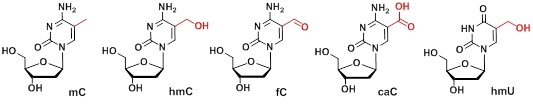
Scheme 1: Depiction of the new epigenetic bases.
[1] M. Wagner, J. Steinbacher, T. F. Kraus, S. Michalakis, B. Hackner, T. Pfaffeneder, A. Perera, M. Müller, A. Giese, H. A. Kretzschmar, T. Carell Angew. Chem. Int. Ed. 2015, doi: 10.1002/anie.201502722. Age-Dependent Levels of 5-Methyl-, 5-Hydroxymethyl-, and 5-Formylcytosine in Human and Mouse Brain Tissues.
[2] Perera, D. Eisen, M. Wagner, S. K. Laube, A. F. Künzel, S. Koch, J. Steinbacher, E. Schulze, V. Splith, N. Mittermeier, M. Müller, M. Biel, T. Carell, S. Michalakis Cell Rep. 2015, 11, 1-12. TET3 Is Recruited by REST for Context-Specific Hydroxymethylation and Induction of Gene Expression
[3] C.G. Spruijt, F. Gnerlich, A.H. Smits, T. Pfaffeneder, P.W.T.C. Jansen, C. Bauer, M. Münzel, M. Wagner, M. Müller, F. Khan, H.C. Eberl, A. Mensinga, A.B. Brinkman, K. Lephikov, U. Müller, J. Walter, R. Boelens, H. van Ingen, H. Leonhardt, T. Carell∗, M. Vermeulen∗ Cell. 2013, 152, 1146-59. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives
[4] S. Becker, I. Thoma, A. Deutsch, T. Gehrke, P. Mayer, H. Zipse, T. Carell, Science, 2016, 352 (6287), 833-836.
A high-yielding, strictly regioselective prebiotic purine nucleoside formation pathway.
 Thomas Carell (Ph. D) was raised in Bad-Salzuflen (Germany). He studied chemistry at the Universities of Münster and Heidelberg. In 1993 he obtained his doctorate with Prof. H. A. Staab at the Max Planck Institute of Medical Research in Heidelberg. After postdoctoral training with Prof. J. Rebek at MIT (Cambridge, USA) in 1993-1995, Thomas Carell moved to the ETH Zürich (Switzerland) as an ass. professor to start independent research. He obtained his habilitation (tenure) in 2000. He subsequently accepted a full professor.
Thomas Carell (Ph. D) was raised in Bad-Salzuflen (Germany). He studied chemistry at the Universities of Münster and Heidelberg. In 1993 he obtained his doctorate with Prof. H. A. Staab at the Max Planck Institute of Medical Research in Heidelberg. After postdoctoral training with Prof. J. Rebek at MIT (Cambridge, USA) in 1993-1995, Thomas Carell moved to the ETH Zürich (Switzerland) as an ass. professor to start independent research. He obtained his habilitation (tenure) in 2000. He subsequently accepted a full professor.
 Funder under FP7 project, The ERA Chair Culture as a Catalyst to Maximize the Potential of CEITEC´ (contract no. 621368).
Funder under FP7 project, The ERA Chair Culture as a Catalyst to Maximize the Potential of CEITEC´ (contract no. 621368).